Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3...
Transcript of Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3...
Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3)4]2 + !
Dibuje los espectros EPR medidos en 300 y 100 K !
1
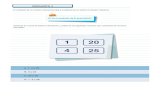
Pregunta: Bajo la aceptación de una geometría cuadrado planar
de Cu(II) dónde es gI (g-perpendicular) y dónde g║(g-parallel) ?
Pregunta : Explica Hyperfine Pattern (alfa protones) de
Naphthalene radical anion ! („Stick diagram“)
Selected Metals: Copper Proteins
Concept of Malkin and Malmström R Malkin, BG Malmström, Advances Enzymology, 33,177-244 (1970)
Classification of Cu sites according to UV/VIS
and EPR Properties: Type 1 (blue; mononuclear),
Type 2 (non-blue; mononuclear), Type 3 (EPR-
silent; dinuclear)
Blumberg-Peisach Plot (gII vs AII) W E Blumberg, J Peisach, in Probes of Structure and Function of Macromolecules and
Membranes, ed. B Chance, Academic Press, New York, 1971, vol. 2, pp. 215–229
J Peisach, WE Blumberg, ARCHIVES OF BIOCHEMISTRY AND BIOPHYSICS, 165, 691-
708 (1974)
2
Blumberg-Peisach Plot - gII vs AII – Cu Proteins
3
Cu A, in Cytochrome c
Oxidase and Nitrous
Oxide Reductase
Bioengineering: tipo 1 Cu → tipo 2 Cu
Handbook of Metalloproteins (A Messerschmidt, T L Poulos, K Wieghardt, R Huber, eds), Wiley, 2001
EPR (a) EPR (c) EPR (d)
Type 1 Cu in
Azurin
Cys112
6
Bioengineering of Electron Transfer Centers : tipo 1 Cu → tipo
CopperA (dinuclear mixed-valent) Work of G Canters/Leiden/NL and Y Lu, Urbana/USA; MG Savelieff, Y Lu, J Biol Inorg Chem, 15,
967-976 (2010)
Plastocyanin/Photosynthesis Cytochrome c Oxidase/Respiration
Nitrous Oxide Reductase/Denitrification 7
Cu Hyperfine Structure – Nitrous Oxide Reductase Coyle, Zumft, Kroneck, Körner, Jakob (1985) Eur. J. Biochem., 153, 459; Neese, Zumft,
Antholine, Kroneck (1996) J. Am. Chem. Soc., 118, 8692; Pomowski, Zumft, Kroneck, Einsle
(2011), Nature, 477, 234
2600 2800 3000 3200 3400
Magnetic Field (G)
CuA ICu=3/2
7-Lines Observed
#Lines=2nI+1
n=2
2x2x3/2+1=7
1:2:3:4:3:2:1
Expected and Observed
2 Equivalent
Cu predicted
8
7-line EPR Spectrum of CuA (2nd derivative)
One electron delocalized over two Cu nuclei
Note the short (metallic) Cu-Cu distance ≈ 2.5 Å
9
Historia 2:El descubrimiento de un nuevo Centro de Fe IDENTIFICATION BY ISOTOPIC SUBSTITUTION OF THE EPR SIGNAL AT g = 1.94 IN A NON-HEME
IRON PROTEIN FROM AZOTOBACTER, Y I SHETHNA, P W WILSON, R E HANSEN, H BEINERT,
Proceedings National Academy of Science/USA (1964), 52, 1263-1271
EPR spectra of Fe56 and
Fe57 iron proteins
superimposed. The dotted
curve represents a
computed curve for the
Fe57 protein, which was
obtained from the curve of
the Fe57 protein, assuming
a hyperfine splitting of 22
G and a final enrichment
of 65% for Fe57 , IFe57 = 1/2.
13
Selected Metals: Iron-Sulfur/FeS Proteins Sulfide (S2-, inorganic/labile) & CyS- (organic/stable)
Spinach [2Fe-2S] Ferredoxin PDB 1A70 Electron Transfer Protein
14
Redox states of [2Fe-2S]-Cluster
ox state mixed-valent state
high-spin Fe high-spin Fe
S = 0 ; diamagnetic S = 1/2 ; paramagnetic; unpaired
e- delocalized over both Fe centers
delocalized
discrete, Fe(II) and Fe(III) centers
•antiferromagnetic coupling between both Fe centers
•[Fe2S2]+ with S = 9/2 (2x Fe(2.5))
REDOX of [2Fe-2S] center
15
Redox states of [4Fe-4S]-Cluster
HiPIP-type Ferredoxin-type
S = ½ S = 0 S = ½
paramagnetic diamagnetic paramagnetic
• Redox potentials: +50 to +450 mV (HiPIP)
-650 to 0 mV (Ferredoxin)
• Higher potentials of HiPIPs: different redox couples
REDOX [4Fe-4S] center antiferromagnetically coupled Fe2S2-units
16
Recordar: Fe-S Centers – EPR Finger Printing
300 320 340 360 380 400
Magnetic field [mT]
[4Fe-4S]3+/2+
HiPIP
[4Fe-4S]2+/+
[2Fe-2S]2+/+ Ferredoxin
[2Fe-2S]2+/+ Rieske
oxidized gav=2.06
[3Fe-4S]+/0
oxidized gav=2.01
reduced gav=1.96
reduced gav=1.96
reduced gav=1.91
gy=1.91
gx=1.79
gz=2.02
gz=2.05
gy=1.95
gx=1.89
gz=2.06 gy=1.92
gx=1.88
gz=2.12 gx,y=2.04
gz=2.02
gx,y=2.00
17
Selected Metals: Molybdenum/Tungsten Proteins
RC Bray, LS Meriwether, Nature, 212, 467-469 (1966)
R Hille, Archives of Biochemistry and Biophysics, 433 107–116 (2005)
Molybdenum EPR signal of Xanthine Oxidase
(XO) reacted with formaldehyde. Bottom trace:
XO, nat. abundance; central trace : Mo-95
enriched XO; upper trace: central trace, enlarged 20
21
????????????????
You figure out the
geometry of the
Mo(V) site !
Hyperfine splittings
from 95Mo and
Hydrogen/Substrate.
EPR Prediction: Acetylene Hydratase is a Tungsten, Iron-Sulfur Enzyme
RU Meckenstock, R Krieger, S Ensign, PMH Kroneck, B Schink, Eur J Biochem, 264, 176–82 (1999)
3300 3400 3500 3600 3700-20
-15
-10
-5
0
5
10
15
20
25
30
A
B
1.920
1.9352.048
Re
lative
in
tensity
Magnetic field [G]
-6
-4
-2
0
2
2,15 2,1 2,05 2 1,95 1,9 1,85
2.005
2.048
C
Rela
tive in
ten
sit
y
2.015
g value
320 330 340 350 360 370 380
Magnetic field [mT]
Substrate
Binding
[4Fe-4S] < 20 K, Electron Transfer
W catalytic site, > 20 K
C2H2 + H2O → [H2C=C(OH)H] → CH3CHO
22
3D Structure of Tungsten Acetylene Hydratase GB Seiffert, MG Ullmann, A Messerschmidt, B Schink, PMH Kroneck,O Einsle, Proc Natl Acad Sci USA,
104, 3073–77 (2007).
PDB code: 2E7Z
23
Tyrosyl Radicals in Biology
Tyrosine:
• Weak O-H Bond (87 kcal/mol) due
to stabilization of the radical
• pKa=10
• Redoxpotential=650-1100 mV ( vs
NHE)
Tyrosyl Rdical Enzymes:
• Ribonucleotide Reductase
• Photosystem II
• Galactose Oxidase
• Catalase
• Prostaglandin H Synthase
24
EPR Spectra of Tyrosyl Radicals
3240 3260 3280 3300 3320 3340
Magnetic Field (G)
87100 87200 87300 87400 87500
2.0091
2.0067
2.0045
2.0023
Magnetic Field (G)
X-Band
9.2 GHz High Field
245 GHz
Large variation in gmax values reflects different protein
environments and carries electronic structure information
E. Coli
RNR
Mouse
RNR
25
H-Bonding Effects on gmax
O
O
gmax
LPop
LPip
• Rotation around the C-O bond direction creates
a large angular momentum along the C-O bond
• gmax is along C-O
• Rotation is the more effective the smaller the
energy gap
• H-bonding quenches angular momentum
1.6 1.7 1.8 1.9 2.0 2.1
8850
8900
8950
9000
9050
9100
9150
H-Bond distance (A)
Exci
tation E
nerg
y (
cm-1)
2.0064
2.0067
2.0070
gm
ax
27
Outlook – EPR técnicas avanzadas
28
S> 1/2 systems: Triplet states ; Transition Metal Ions (e.g. h.s. Fe(III) and
Fe(II); Heme Fe; Mn(II), Mn(III), Mn(IV))
Wertz, Bolton (1986) Electron Spin Resonance, Elementary Theory and Practical
Applications, Chapmann and Hall; Cammack, Cooper (1993) Meth. Enzymol.
227, 353.
Advanced techniques: ENDOR; Pulse EPR/ENDOR; Spin Labeling;
Distance Measurements; Imaging
Mims, Proc. R. Soc. Lond. A (1965) 283, 452; Hoffman (2003) PNAS 100, 3575;
Murphy, Farley (2006) Chem. Soc. Rev. 35, 249; Eaton, Eaton (2012) J. Magn.
Reson., 223,151; Van Doorslaer, Desmet (2008) Meth. Enzymol. 437, 287;
Roser, Schmidt, Drescher, Summerer (2016) Org. Biomol. Chem. 14, 5468.
Adenosine-5‘-phosphate Reductase (PDB code: 1JNR) G Fritz, A Roth, A Schiffer, T Büchert, K Bourenkov, HD Bartunik, H Huber, KO Stetter, PMH
Kroneck, U Ermler, Proc Natl Acad Sci USA, 99, 1836-1841 (2002)
29
Interaction between the two [4Fe-4S] - Centers
G Fritz, T Büchert, PMH Kroneck, J Biol Chem, 277, 26066-26073 (2002)
10.8 Ǻ
310 320 330 340 350 360 370 380
Magnetic field [mT]
2.078 1.941 1.902 2.066 2.045 1.976 1.938 1.890
310 320 330 340 350 360 370 380
Magnetic field [mT]
I II I II
Eo‘ = - 60 mV Eo‘ = - 520 mV
[+] [2+] [+] [+]
15.4 Å9.8 Å
R
II I
30
ENDOR (Electron Nuclear DOuble Resonance) provides missing information
3390 3400 3410 3420
g = 2.0026
Magnetic Field / Gauss
8 10 12 14 16 18 20 22
Mutant
12I
12II2II
7II
7I
2I
1'
7II2II
2I
12I
12II
17
17
18
18
7I2'
2
Frequency / MHz
1
WT
H
P700 radical cation
(Chlorophyll a dimer)
EPR
ENDOR
31
Applied for many biological systems with small hyperfine coupling
constants which are not resolved in conventional CW EPR spectrum
3360 3370 3380 3390 3400 3410
Magnetic Field / Gauss
2 4 6 8 10 12 14 16 18 20 22 24 26 28
H = 14.3 MHz
Frequency / MHz
.1
2
NMR = n a/2
A1
A2
EPR ENDOR
Phenalenyl radical
Hyperfine enhancement effect
32
Spin labeling- Peptides and Proteins
R N
O
OH
N
HO
O
+DCC O
NO
R
OO
N O
Nitroxides are introduced into proteins as reporter groups to provide information
about local environment, overall tumbling rate of the protein or/and segmental
mobility, accessibility of the labeling site for polar/non-polar molecules, distance
measurements to other spin labels, co-factors, membrane surface….
Labeling of the hydroxyl group
NO
CH2SSO2CH3Protein SH + S CH2
NO
SProtein
MTSL spin label is cysteine specific.
SDSL = site directed spin labeling is introducing cysteines into the protein
molecule by point mutations with following MTSL labeling.
Cysteine mapping of the protein.
33
Modern pulse EPR techniques (DQC, DEER) can cancel out all interactions
resulting in an EPR spectrum except the dipole interaction in spin pairs
Example: spin labeled Gramicidin A
3/2 re D
dipolar, MHz,
2/][]Å[
102.53
4
MHzr
dipolarD
Å.930Interspin distance=
34 DEER = double electron electron resonance; DQC = double-quantum coherence
DQC-EPR ruler
G-C
C-G
A-U
G-C
C-G
U-A
G-C
A-U
U-A
G-C
G-C
C-G
C-G
U-A
A-U
C-G
G-C
C-G
G-C
U-A
G-C
U-A
U-A
C-G
(O*N) -U
5'
3'
U-(N*O)3'
5'
G-C
C-G
A-U
G-C
C-G
U-A
G-C
A-U
U-A
G-C
G-C
C-G
C-G
U-A
A-U
C-G
G-C
C-G
G-C
U-A
G-C
U-A
U-A
C-G
(O*N) -U
5'
3'
U-(N*O)3'
5'
10 20 7030 40 50 60 1009080
DQC RULER
Å
Location of mobile domains in a protein
complex using DQC-EPR
35
Oxygen Accessibility
W. L. Hubbell, H. S. Mchaourab, C. Altenbach, and M. A. Lietzow,
Structure 4, 779-783 (1996).
Oxygen accessibility and probe
mobility were measured as a
function of sequence number for
spin labels attached to T4
lysozyme (T4L) and cellular
retinol binding protein (CRBP).
The correlation between the two
parameters indicates that the
most mobile sites are also the
most oxygen accessible.
The repeat period of about 3.6
for T4L is consistent with the a-
helical structure of this segment
of the protein.
36
EPRI - See Video Center for EPR Imaging, The University of Chicago, USA
https://epri.uchicago.edu/page/epr-imaging
37
![Page 1: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/1.jpg)
![Page 2: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/2.jpg)
![Page 3: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/3.jpg)
![Page 4: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/4.jpg)
![Page 5: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/5.jpg)
![Page 6: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/6.jpg)
![Page 7: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/7.jpg)
![Page 8: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/8.jpg)
![Page 9: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/9.jpg)
![Page 10: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/10.jpg)
![Page 11: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/11.jpg)
![Page 12: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/12.jpg)
![Page 13: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/13.jpg)
![Page 14: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/14.jpg)
![Page 15: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/15.jpg)
![Page 16: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/16.jpg)
![Page 17: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/17.jpg)
![Page 18: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/18.jpg)
![Page 19: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/19.jpg)
![Page 20: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/20.jpg)
![Page 21: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/21.jpg)
![Page 22: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/22.jpg)
![Page 23: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/23.jpg)
![Page 24: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/24.jpg)
![Page 25: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/25.jpg)
![Page 26: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/26.jpg)
![Page 27: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/27.jpg)
![Page 28: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/28.jpg)
![Page 29: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/29.jpg)
![Page 30: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/30.jpg)
![Page 31: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/31.jpg)
![Page 32: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/32.jpg)
![Page 33: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/33.jpg)
![Page 34: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/34.jpg)
![Page 35: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/35.jpg)
![Page 36: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/36.jpg)
![Page 37: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/37.jpg)
![Page 38: Pregunta : Tiene una solución de 1.0 mM de [Cu(NH3 4depa.fquim.unam.mx/amyd/archivero/BIOEPRUNAM_03112016_01_33366.pdf · Pregunta : Tiene una solución de 1.0 mM de [Cu(NH 3) 4]2](https://reader039.fdocuments.es/reader039/viewer/2022040217/5d64a91f88c993952a8b81d7/html5/thumbnails/38.jpg)